|
Cardiovascular Biomechanics and A.I. Laboratory |
|
|
Imperial College London, Department of bioengineering |
|
Machine
Learning Biomechanics |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Overall
Methodology We
employ deep learning networks that are constrained by physics governing
equations to extract motion, myocardial strains, and to perform computational
flow simulations as well as finite element myocardial tissue mechanics
simulations. Combined with our deep learning image processing work, we aim to
have pipelines that start directly from the images for seamless processing. Multi-Case
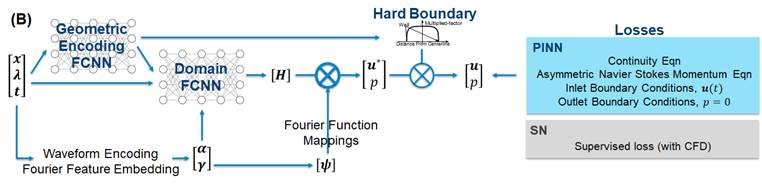
PINN for Vascular Flow Simulations We
developed a neural network pre-trained over a wide range of possible vascular
geometries, by parameterizing the geometries as network inputs. This will
enable flow simulation results to be very quickly generated when a new case
that is unseen by the network is presented. We compared PINN to a network
directly supervised by flow simulation results (SN), but find that SN
outperforms PINN in this problem setup, both in terms of accuracy and
computational cost. We further discovered that some auxiliary strategies can
enhance performance, such as geometric encoding (a pre-trained network that
calculates curvilinear coordinate parameters), and hard constrained no-slip
boundary condition. [We
are in the process of publishing this work to provide further details]
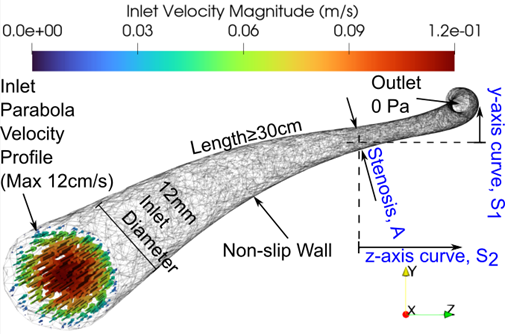
Problem
definition: to predict velocities and pressures in a curved stenotic vessel,
whose curvatures and stenosis severity are within a range of possible values.
Flow can be steady or pulsatile.
Our network architecture
Errors of network prediction of velocities and
pressure in 279 unseen testing geometries within the trained geometric parameter
range, expressed as the average or maximum of all testing geometries (where
the spatial maximum error is taken as the error of each geometry). |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
PINN
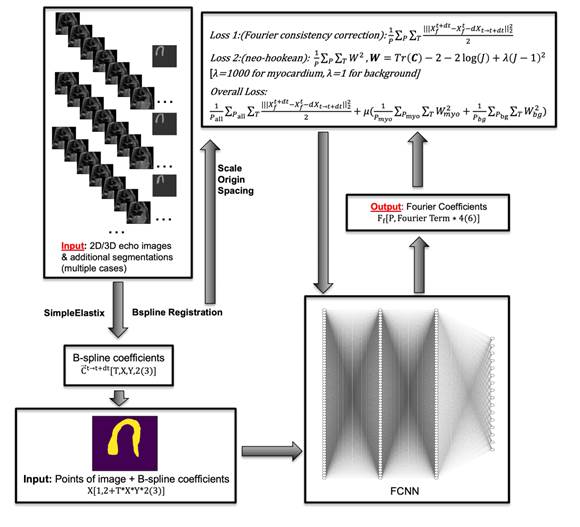
Image Registration of MRI Images We
developed a new Physics Informed Neural Network (PINN) framework tailored for
left ventricular (LV) finite element (FE) modelling. Compared to existing
PINN-FE approaches, our approach introduces 2 key innovations: (1)
Consistency with imaged motions: our PINN enforces an alignment between
simulated cardiac motions and image-derived motion, so that predictions will
have a high level of fidelity to imaged cardiac behaviour. (2) Estimations of
myocardial stiffness and active tension: our PINN framework performs a
back-computation of these parameters, to reach patient-specificity. Our
PINN-FE utilizes imaged shape modes to speed up computations, and requires
only 3 minutes for training each case. This gives it a speed advantage over
traditional FE and the alternative PINN approaches. [We
are in the process of publishing this work to provide further details]
Workflow
of our FINN-FE framework |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
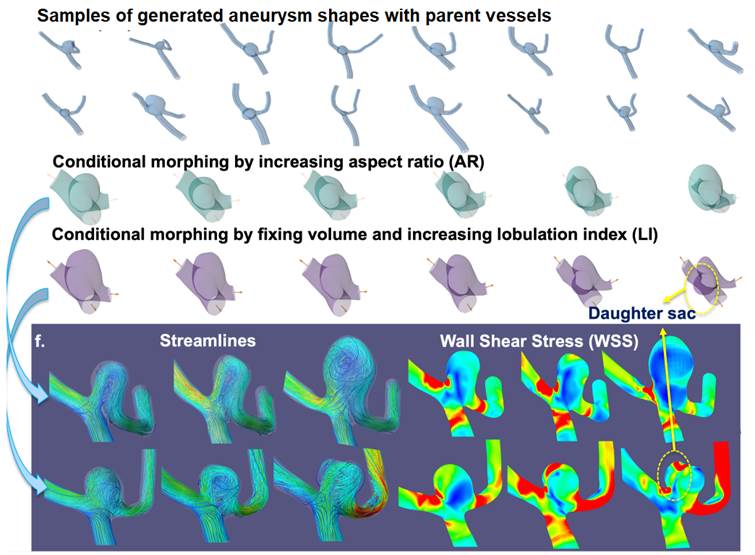
Deep
Learning Generation of Realistic Cranial Aneurysm Geometries Fluid
mechanical stresses are believed to play an important role in determining the
rupture and disease progression risks of cranial aneurysm. Performing
image-based simulations to obtain such fluid dynamics details is time
consuming. We aspire to train a network for real time prediction of fluid
dynamics parameters for clinical uses. To do this, we first need to resolve
the lack of a large dataset of cranial aneurysms and its computational fluid
dynamics (CFD) simulations. We developed a generative AI network to generate
realistic synthetic cranial aneurysm 3D meshes, which is composed of both the
parent vessels and the aneurysm pouch, which is ready for CFD simulations. We
utilize Graph Fourier Deformation to model shapes, and utilized variable
autoencoders for the generation. A
particular useful feature of our shape generator is that it can be controlled
to generate specific clinically relevant shape parameters, such as specific
aneurysm pouch aspect ratio, neck size, and extent of lobulation. This is
useful for downstream fluid dynamics investigations to understand effects of
geometric features on fluid patterns and stresses. [We
are in the process of publishing this work to provide further details]
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||